-Search query
-Search result
Showing 1 - 50 of 89 items for (author: hsu & std)

EMDB-38650: 
Additional map for SARS-CoV-2 Spike D614G variant, one RBD-up conformation 1 (PDB ID: 7EAZ; EMD-31047). Map was generated from heterogeneous refinement with downsampling in CryoSPARC
Method: single particle / : Yang TJ, Yu PY, Hsu STD

EMDB-33942: 
Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33943: 
Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 1
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33944: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33945: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 3
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33946: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33947: 
Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33948: 
Cryo-EM structure of MERS-CoV spike protein, intermediate conformation
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-33949: 
Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P

EMDB-31820: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Kappa variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD
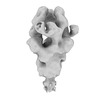
EMDB-31818: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Gamma variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31822: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Kappa variant in complex with neutralizing antibody RBD-chAb-25
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31821: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Delta variant in complex with neutralizing antibody RBD-chAb-25
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31817: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Beta variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31819: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Delta variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD
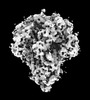
EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
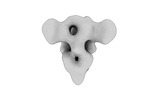
EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
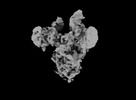
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
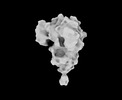
EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33702: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
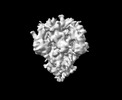
EMDB-33703: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33704: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
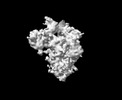
EMDB-33705: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
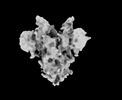
EMDB-33706: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-31760: 
Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, one RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31761: 
Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, two RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31762: 
Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31763: 
Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31764: 
Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), two RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31767: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), all RBD-down conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31768: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31769: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31770: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), two RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31771: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31772: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31773: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31775: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), all RBD-down conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31776: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31777: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31778: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 3
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31779: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
